Difference between revisions of "Coupling with WRF-Chem"
(→Running an idealized example: adding Matlab line from Jon) |
|||
| (53 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{users guide}} | {{users guide}} | ||
| − | As of WRF | + | As of WRF 4, WRF-Chem is now included in the WRF repository. Fire emissions can now input into WRF-Chem as chemical species or a simple smoke tracer. [[File:Sfire smoke.png|thumb|SFire smoke tracer in action. See [[File:Smoke small.avi|animation]].]] |
| − | WRF-Chem | ||
| − | |||
| − | [[File:Sfire smoke.png|thumb|SFire smoke tracer in action. See [[File:Smoke small.avi|animation]].]] | ||
==Compiling with chem support== | ==Compiling with chem support== | ||
Compiling the code with support for WRF-Chem is similar to the [[How to compile WRF-Fire|standard procedure]]. | Compiling the code with support for WRF-Chem is similar to the [[How to compile WRF-Fire|standard procedure]]. | ||
| − | You must set an environment variable | + | You must set an environment variable '''WRF_CHEM''' and configure with the argument '''chem''' as follows. |
<pre> | <pre> | ||
export WRF_CHEM=1 | export WRF_CHEM=1 | ||
| Line 16: | Line 13: | ||
==Running an idealized example== | ==Running an idealized example== | ||
| − | An idealized example based on the hill case has been created in | + | An idealized example based on the hill case has been created in [https://github.com/jbeezley/wrf-fire/tree/master/wrfv2_fire/test/em_fire/chem '''test/em_fire/chem'''] |
| − | + | Note that '''sfc_full_init =.true.''' is required in the '''&fire''' section of '''namelist.input''' to initialize USGS landuse, which is required by WRF-Chem. | |
| − | + | The tracer selected by '''tracer_opt=1''' will appear as an atmospheric variable in the output file with the name '''smoke'''. The chemical species in the atmosphere, selected by '''chem_opt''', will appear under their respective names, such as '''n2o5'''. | |
| − | WRF | + | |
| − | + | ==Configuration== | |
| − | + | The selection of the model in WRF-Chem is controlled by namelist variables '''tracer_opt''' in the [https://github.com/jbeezley/wrf-fire/blob/4c0851085eeb25d9143bd5b07e736fe949700497/wrfv2_fire/test/em_fire/chem/namelist.input#L78 '''&dynamics'''] section and '''chem_opt''' in the [https://github.com/jbeezley/wrf-fire/blob/4c0851085eeb25d9143bd5b07e736fe949700497/wrfv2_fire/test/em_fire/chem/namelist.input#L203 '''&chemistry'''] section of '''namelist.input'''. At the moment, the coupling supports '''tracer_opt = 1''', which creates a single tracer called '''smoke''', and a subset of RADM2 ('''chem_opt=2'''), MOZART ('''chem_opt=112''' or '''119'''), and GOCART ('''chem_opt=300''') variables. | |
| − | + | ||
| − | + | The option '''tracer_opt = 2''' does not require WRF-Chem, and it creates passive tracers called '''tr17_1''' to '''tr17_8'''. Emissions \are inserted proportionally to the fuel burned according to the emission factors in file '''namelist.fire_emissions'''. Properties of the tracers '''tr17_1''' to '''tr17_4''' are identical, but the others slightly differ. | |
| − | + | ||
| + | To use the passive tracers in the code built with WRF-Chem, specify '''tracer_opt=2''' and '''chem_opt=0''' in '''namelist.input'''. The tracers work slightly differently in code built with and without WRF-Chem, as described in file '''chem/module_input_tracer.F'''. A sample emission factors table for this usage is available as '''test/em_fire/chem/namelist.fire_emissions.tracers'''. It can be used in ideal as well as real runs. Different rows in the table can be used to track separate species, e.g., pm10 and pm25, with different emission factors. | ||
| + | |||
| + | In each time step, the coupling code adds the fire emissions created in the time step to the first layer of atmosphere cells. The chemistry state arrays are have unit of concentrations ('''ppmv'''), so the amounts of the inserted species are scaled by the dry air mass in the first layer. The amount of the chemical species created is determined from the amount of fuel burned directly by an emissions table, while the smoke tracer uses the fire ground heat flux '''GRNHFX''' as a proxy. The emissions table contains the amounts of emissions fuel burned in '''g/kg''' or '''mol/kg'''. The emissions tables are specified in file '''namelist.fire_emissions'''. This file contains a line for each species like | ||
| + | <pre> | ||
| + | no2=3.2,3.2,3.2,1.4,1.4,1.4,1.4,2.7,2.7,2.7,3,3,3, | ||
| + | </pre> | ||
| + | with emissions (g/kg fuel or mol/kg fuel) for each of the fuel categories used (by default, the 13 Anderson's categories). | ||
| + | |||
| + | There are two additional variables in '''namelist.fire_emissions''': | ||
| + | <pre> | ||
| + | compatible_chem_opt=112,119 | ||
| + | </pre> | ||
| + | is a list of '''chem_opt''' values intended to be used with the table. However, presently, the code does not check if the file lists other chemistry species supported by the coupling code, not just those for the given '''chem_opt'''. If it does, the results can be upredictable. | ||
| + | The variable | ||
<pre> | <pre> | ||
| − | + | printsums=1 | |
| − | |||
| − | |||
</pre> | </pre> | ||
| − | + | will enable printing of the totals of each emissions species at the end of every time step. | |
| + | |||
| + | Several prototype emissions tables are provided in '''test/em_fire/chem'''. You need to copy or link one of the emissions tables to '''namelist.fire_emissions''' in your run directory, such as | ||
<pre> | <pre> | ||
| − | + | ln -s namelist.fire_emissions.RADM2 namelist.fire_emissions | |
</pre> | </pre> | ||
| − | + | and set matching '''chem_opt''' in '''namelist.input''', here '''chem_opt=2'''. See the next section for the available emissions tables. Users can also add their own. | |
| − | == | + | ==Chemical species== |
| − | + | ||
| − | the | + | The chemical model selected by the '''chem_opt''' flag produces concentrations on 3D grids in every '''wrfout''' file produced, for a set of variables defined in the '''Chem scalars''' part of the file [https://github.com/openwfm/WRF-SFIRE/blob/262829290d98e419b6839893ecb1a5d36ed4b969/Registry/registry.chem#L1567 '''Registry/registry.chem''']. The first section of the list contains the chemical species common to all models, later sections list species added for specific models. For more detailed information about the species including dusts and aerosols in specific WRF-Chem models, see the [https://ruc.noaa.gov/wrf/wrf-chem/model_info.htm WRF-Chem model documention] and the links to the individual models at the emissions tables below. |
| − | + | ||
| − | + | The quantities of the chemical species are in '''ppmv''', i.e., 1e6*mol/mol of dry air. | |
| − | + | ||
| + | The frequency of writing the outputs into the wrfout files is specified by the user in the '''history_interval''' flags in the WRF '''namelist.input''' file. It can be as often as once every minute, but usually much less. | ||
| + | |||
| + | SFIRE adds the fire emissions to a subset of the chemical species as specified in the emissions table. It is up to the chemical model to produce other species from them. Note that the parts of the registry.chem file using the word "emissions" are for other emission schemes and not relevant here. SFIRE adds fire emissions directly to the lowest layer of the 3D Chem fiels during the fire simulation. | ||
| + | |||
| + | There are currently three emissions tables available: | ||
| + | * [https://github.com/openwfm/WRF-SFIRE/blob/master/test/em_fire/chem/namelist.fire_emissions.RADM2 namelist.fire_emissions.RADM2] for the [http://ruc.noaa.gov/wrf/WG11/radm2.htm RADM2] model | ||
| + | * [https://github.com/openwfm/WRF-SFIRE/blob/master/test/em_fire/chem/namelist.fire_emissions.MOZART namelist.fire_emissions.MOZART] for the [http://www.acd.ucar.edu/wrf-chem/mozart.shtml MOZART] model | ||
| + | * [https://github.com/openwfm/WRF-SFIRE/blob/master/test/em_fire/chem/namelist.fire_emissions.GOCART namelist.fire_emissions.GOCART] for the [https://tropo.gsfc.nasa.gov/gocart GOCART] model | ||
| + | |||
| + | ==Implementation== | ||
WRF-Chem contains a multitude of arrays representing concentrations of chemical species. The typical | WRF-Chem contains a multitude of arrays representing concentrations of chemical species. The typical | ||
method of injecting data into the simulation involves generating emission input data files from standard | method of injecting data into the simulation involves generating emission input data files from standard | ||
sources. These sources are read into the code using auxiliary input streams and interpolated into | sources. These sources are read into the code using auxiliary input streams and interpolated into | ||
| − | the species arrays in the | + | the species arrays in the [https://github.com/jbeezley/wrf-fire/blob/4c0851085eeb25d9143bd5b07e736fe949700497/wrfv2_fire/chem/module_emissions_anthropogenics.F emissions] module. For the coupling with SFIRE, turning off file |
input in the namelist seems to initialize the emission arrays to zero. This eliminates the need to generate | input in the namelist seems to initialize the emission arrays to zero. This eliminates the need to generate | ||
| − | useless emission files. | + | useless emission files. |
| + | |||
| + | WRF-Chem stores its state in arrays '''chem''' and '''tracer'''. These arrays are four dimensional indexed like '''(x,z,y,s)''', where s is the chemical | ||
| + | species. The domain structure contains scalars such as '''p_smoke''' (into the '''tracer''' array) and '''p_nh4''' (into the '''chem''' array) that give the species index for | ||
| + | each type. The species selections are specific to different chemistry options ('''chem_opt''') | ||
| + | from the registry; however, even unused species have their index set to 1. The dynamics section of the namelist has an option for '''tracer_opt'''. This | ||
| + | controls what tracers are generated for Chem. | ||
| + | |||
| + | Since fuel is given at the fire grid resolution, the emissions are computed at the fire resolution and then averaged to the atmospheric resolution, where the chemistry operates. However, since the memory demands of a 3D array at fire resolution (two surface dimensions and chemical species) would be excessive, the contributions of the emissions are computed on the fly and added to the respective atmospheric cell immediately. This results in code that straddles the otherwise strict division in SFIRE between fire computations on the fire grid, which are independent of WRF, and the atmospheric computations related to WRF, which otherwise consist of interpolation only and should not contain any other code. So, subroutine [https://github.com/jbeezley/wrf-fire/blob/4c0851085eeb25d9143bd5b07e736fe949700497/wrfv2_fire/phys/module_fr_sfire_atm.F#L276 '''add_fire_emissions_to_chem'''], which adds chemical species to the '''chem''' array, was derived from the utility [https://github.com/jbeezley/wrf-fire/blob/4c0851085eeb25d9143bd5b07e736fe949700497/wrfv2_fire/phys/module_fr_sfire_util.F#L471 '''sum_2d_cells'''] by adding chemistry-specific code. | ||
| − | WRF-Chem | + | The chemistry coupling code is compiled only when WRF-Chem is being compiled. It resides in [https://github.com/jbeezley/wrf-fire/blob4c0851085eeb25d9143bd5b07e736fe949700497/wrfv2_fire/phys/module_fr_sfire_atm.F module_fr_sfire_atm.F], which is allowed by the SFIRE architecture to contain code both at atmosphere and fire resolution. Besides subroutine '''add_fire_emissions_to_chem''', the code consists of subroutine [https://github.com/jbeezley/wrf-fire/blob/4c0851085eeb25d9143bd5b07e736fe949700497/wrfv2_fire/phys/module_fr_sfire_atm.F#L151 '''read_emissions_table'''], which reads file '''namelist.fire_emissions''', subroutine [https://github.com/jbeezley/wrf-fire/blob/4c0851085eeb25d9143bd5b07e736fe949700497/wrfv2_fire/phys/module_fr_sfire_atm.F#L276 '''add_fire_emissions_to_chem'''], which adds chemical species to the '''chem''' array, the [https://github.com/jbeezley/wrf-fire/blob/4c0851085eeb25d9143bd5b07e736fe949700497/wrfv2_fire/phys/module_fr_sfire_atm.F#L78 emissions tables] as module variables, subroutine [https://github.com/jbeezley/wrf-fire/blob/4c0851085eeb25d9143bd5b07e736fe949700497/wrfv2_fire/phys/module_fr_sfire_atm.F#L1914 '''fire_emission'''], which adds the smoke tracer, and the corresponding call statements in [https://github.com/jbeezley/wrf-fire/blob/4c0851085eeb25d9143bd5b07e736fe949700497/wrfv2_fire/phys/module_fr_sfire_driver.F '''module_fr_sfire_driver''']. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | tracer | ||
| − | + | ==References== | |
| − | + | * Kochanski A. K., Jenkins M.A., Yedinak K., Mandel J., Beezley J, and Lamb B. (2015) '''Toward an integrated system for fire, smoke and air quality simulations''', International Journal of Wildland Fire - http://dx.doi.org/10.1071/WF14074 | |
| − | + | * Adam K. Kochanski, Jonathan D. Beezley, Jan Mandel, Craig B. Clements, '''Air pollution forecasting by coupled atmosphere-fire model WRF and SFIRE with WRF-Chem''', 4th Fire Behavior and Fuels Conference, February 18-22, 2013, Raleigh, NC, {{arXiv|1304.7703}}, 2013 | |
| − | + | * Adam K. Kochanski, Jonathan D. Beezley, Jan Mandel, and Minjeong Kim, '''WRF fire simulation coupled with a fuel moisture model and smoke transport by WRF-Chem''', [http://www.mmm.ucar.edu/events/2012_wrfusers/ 2012 WRF Users Workshop], [http://www.openwfm.org/wiki/File:WRF_workshop_2012_poster.pdf Poster 51] [http://www.mmm.ucar.edu/events/2012_wrfusers/abstracts/p51.htm abstract] [https://www.regonline.com/AttendeeDocuments/1077122/43365086/wrf2012.pdf paper] {{arXiv|1208.1059}} | |
| + | * [http://ruc.noaa.gov/wrf/WG11/Users_guide.pdf WRF-Chem Users' Guide] | ||
| + | * Georg A. Grell, Steven E. Peckham, Rainer Schmitz, Stuart A. McKeen, Gregory Frost, William C. Skamarock, Brian Eder, [http://dx.doi.org/10.1016/j.atmosenv.2005.04.027 Fully coupled “online” chemistry within the WRF model], Atmospheric Environment, Volume 39, Issue 37, December 2005, Pages 6957–6975 | ||
| + | * C. Wiedinmyer, S. K. Akagi, R. J. Yokelson, L. K. Emmons, J. A. Al-Saadi, J. J. Orlando, and A. J. Soja. [http://dx.doi.org/10.5194/gmd-4-625-2011 The Fire INventory from NCAR (FINN): a high resolution global model to estimate the emissions from open burning], Geosci. Model Dev., 4, 625-641, 2011 | ||
| + | * L. K. Emmons, S. Walters, P. G. Hess, J.-F. Lamarque, G. G. Pfister, D. Fillmore, C. Granier, A. Guenther, D. Kinnison, T. Laepple, J. Orlando, X. Tie, G. Tyndall, C. Wiedinmyer, S. L. Baughcum, and S. Kloster, [http://dx.doi.org/10.5194/gmd-3-43-2010 Description and evaluation of the Model for Ozone and Related chemical Tracers, version 4 (MOZART-4)], Geosci. Model Dev., 3, 43-67, 2010 | ||
| − | + | ==External links== | |
| − | + | * [http://ruc.noaa.gov/wrf/WG11/ WRF-Chem] home page | |
| + | * [https://www2.acom.ucar.edu/modeling/finn-fire-inventory-ncar Finn] emissions model | ||
| + | * [http://www.acd.ucar.edu/wrf-chem/mozart.shtml MOZART] chemistry model | ||
Latest revision as of 04:50, 10 February 2024
- Back to the WRF-SFIRE user guide.
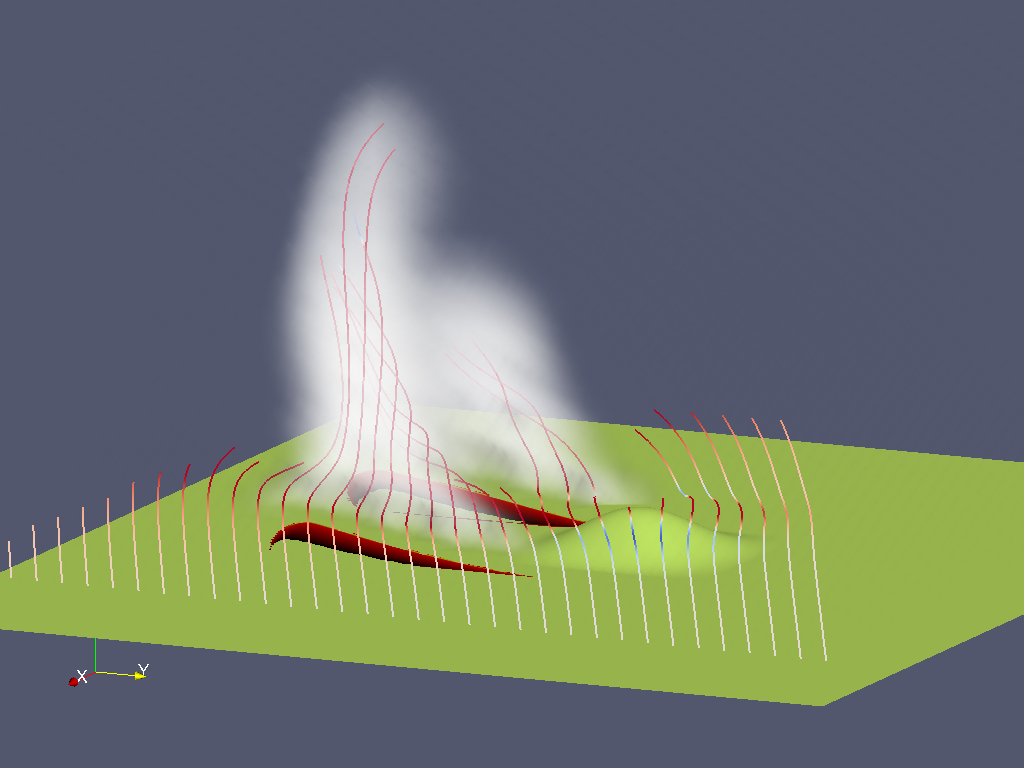
As of WRF 4, WRF-Chem is now included in the WRF repository. Fire emissions can now input into WRF-Chem as chemical species or a simple smoke tracer.
Compiling with chem support
Compiling the code with support for WRF-Chem is similar to the standard procedure. You must set an environment variable WRF_CHEM and configure with the argument chem as follows.
export WRF_CHEM=1 ./configure chem ./compile em_fire
Running an idealized example
An idealized example based on the hill case has been created in test/em_fire/chem Note that sfc_full_init =.true. is required in the &fire section of namelist.input to initialize USGS landuse, which is required by WRF-Chem. The tracer selected by tracer_opt=1 will appear as an atmospheric variable in the output file with the name smoke. The chemical species in the atmosphere, selected by chem_opt, will appear under their respective names, such as n2o5.
Configuration
The selection of the model in WRF-Chem is controlled by namelist variables tracer_opt in the &dynamics section and chem_opt in the &chemistry section of namelist.input. At the moment, the coupling supports tracer_opt = 1, which creates a single tracer called smoke, and a subset of RADM2 (chem_opt=2), MOZART (chem_opt=112 or 119), and GOCART (chem_opt=300) variables.
The option tracer_opt = 2 does not require WRF-Chem, and it creates passive tracers called tr17_1 to tr17_8. Emissions \are inserted proportionally to the fuel burned according to the emission factors in file namelist.fire_emissions. Properties of the tracers tr17_1 to tr17_4 are identical, but the others slightly differ.
To use the passive tracers in the code built with WRF-Chem, specify tracer_opt=2 and chem_opt=0 in namelist.input. The tracers work slightly differently in code built with and without WRF-Chem, as described in file chem/module_input_tracer.F. A sample emission factors table for this usage is available as test/em_fire/chem/namelist.fire_emissions.tracers. It can be used in ideal as well as real runs. Different rows in the table can be used to track separate species, e.g., pm10 and pm25, with different emission factors.
In each time step, the coupling code adds the fire emissions created in the time step to the first layer of atmosphere cells. The chemistry state arrays are have unit of concentrations (ppmv), so the amounts of the inserted species are scaled by the dry air mass in the first layer. The amount of the chemical species created is determined from the amount of fuel burned directly by an emissions table, while the smoke tracer uses the fire ground heat flux GRNHFX as a proxy. The emissions table contains the amounts of emissions fuel burned in g/kg or mol/kg. The emissions tables are specified in file namelist.fire_emissions. This file contains a line for each species like
no2=3.2,3.2,3.2,1.4,1.4,1.4,1.4,2.7,2.7,2.7,3,3,3,
with emissions (g/kg fuel or mol/kg fuel) for each of the fuel categories used (by default, the 13 Anderson's categories).
There are two additional variables in namelist.fire_emissions:
compatible_chem_opt=112,119
is a list of chem_opt values intended to be used with the table. However, presently, the code does not check if the file lists other chemistry species supported by the coupling code, not just those for the given chem_opt. If it does, the results can be upredictable. The variable
printsums=1
will enable printing of the totals of each emissions species at the end of every time step.
Several prototype emissions tables are provided in test/em_fire/chem. You need to copy or link one of the emissions tables to namelist.fire_emissions in your run directory, such as
ln -s namelist.fire_emissions.RADM2 namelist.fire_emissions
and set matching chem_opt in namelist.input, here chem_opt=2. See the next section for the available emissions tables. Users can also add their own.
Chemical species
The chemical model selected by the chem_opt flag produces concentrations on 3D grids in every wrfout file produced, for a set of variables defined in the Chem scalars part of the file Registry/registry.chem. The first section of the list contains the chemical species common to all models, later sections list species added for specific models. For more detailed information about the species including dusts and aerosols in specific WRF-Chem models, see the WRF-Chem model documention and the links to the individual models at the emissions tables below.
The quantities of the chemical species are in ppmv, i.e., 1e6*mol/mol of dry air.
The frequency of writing the outputs into the wrfout files is specified by the user in the history_interval flags in the WRF namelist.input file. It can be as often as once every minute, but usually much less.
SFIRE adds the fire emissions to a subset of the chemical species as specified in the emissions table. It is up to the chemical model to produce other species from them. Note that the parts of the registry.chem file using the word "emissions" are for other emission schemes and not relevant here. SFIRE adds fire emissions directly to the lowest layer of the 3D Chem fiels during the fire simulation.
There are currently three emissions tables available:
- namelist.fire_emissions.RADM2 for the RADM2 model
- namelist.fire_emissions.MOZART for the MOZART model
- namelist.fire_emissions.GOCART for the GOCART model
Implementation
WRF-Chem contains a multitude of arrays representing concentrations of chemical species. The typical method of injecting data into the simulation involves generating emission input data files from standard sources. These sources are read into the code using auxiliary input streams and interpolated into the species arrays in the emissions module. For the coupling with SFIRE, turning off file input in the namelist seems to initialize the emission arrays to zero. This eliminates the need to generate useless emission files.
WRF-Chem stores its state in arrays chem and tracer. These arrays are four dimensional indexed like (x,z,y,s), where s is the chemical species. The domain structure contains scalars such as p_smoke (into the tracer array) and p_nh4 (into the chem array) that give the species index for each type. The species selections are specific to different chemistry options (chem_opt) from the registry; however, even unused species have their index set to 1. The dynamics section of the namelist has an option for tracer_opt. This controls what tracers are generated for Chem.
Since fuel is given at the fire grid resolution, the emissions are computed at the fire resolution and then averaged to the atmospheric resolution, where the chemistry operates. However, since the memory demands of a 3D array at fire resolution (two surface dimensions and chemical species) would be excessive, the contributions of the emissions are computed on the fly and added to the respective atmospheric cell immediately. This results in code that straddles the otherwise strict division in SFIRE between fire computations on the fire grid, which are independent of WRF, and the atmospheric computations related to WRF, which otherwise consist of interpolation only and should not contain any other code. So, subroutine add_fire_emissions_to_chem, which adds chemical species to the chem array, was derived from the utility sum_2d_cells by adding chemistry-specific code.
The chemistry coupling code is compiled only when WRF-Chem is being compiled. It resides in module_fr_sfire_atm.F, which is allowed by the SFIRE architecture to contain code both at atmosphere and fire resolution. Besides subroutine add_fire_emissions_to_chem, the code consists of subroutine read_emissions_table, which reads file namelist.fire_emissions, subroutine add_fire_emissions_to_chem, which adds chemical species to the chem array, the emissions tables as module variables, subroutine fire_emission, which adds the smoke tracer, and the corresponding call statements in module_fr_sfire_driver.
References
- Kochanski A. K., Jenkins M.A., Yedinak K., Mandel J., Beezley J, and Lamb B. (2015) Toward an integrated system for fire, smoke and air quality simulations, International Journal of Wildland Fire - http://dx.doi.org/10.1071/WF14074
- Adam K. Kochanski, Jonathan D. Beezley, Jan Mandel, Craig B. Clements, Air pollution forecasting by coupled atmosphere-fire model WRF and SFIRE with WRF-Chem, 4th Fire Behavior and Fuels Conference, February 18-22, 2013, Raleigh, NC, arXiv:1304.7703, 2013
- Adam K. Kochanski, Jonathan D. Beezley, Jan Mandel, and Minjeong Kim, WRF fire simulation coupled with a fuel moisture model and smoke transport by WRF-Chem, 2012 WRF Users Workshop, Poster 51 abstract paper arXiv:1208.1059
- WRF-Chem Users' Guide
- Georg A. Grell, Steven E. Peckham, Rainer Schmitz, Stuart A. McKeen, Gregory Frost, William C. Skamarock, Brian Eder, Fully coupled “online” chemistry within the WRF model, Atmospheric Environment, Volume 39, Issue 37, December 2005, Pages 6957–6975
- C. Wiedinmyer, S. K. Akagi, R. J. Yokelson, L. K. Emmons, J. A. Al-Saadi, J. J. Orlando, and A. J. Soja. The Fire INventory from NCAR (FINN): a high resolution global model to estimate the emissions from open burning, Geosci. Model Dev., 4, 625-641, 2011
- L. K. Emmons, S. Walters, P. G. Hess, J.-F. Lamarque, G. G. Pfister, D. Fillmore, C. Granier, A. Guenther, D. Kinnison, T. Laepple, J. Orlando, X. Tie, G. Tyndall, C. Wiedinmyer, S. L. Baughcum, and S. Kloster, Description and evaluation of the Model for Ozone and Related chemical Tracers, version 4 (MOZART-4), Geosci. Model Dev., 3, 43-67, 2010